https://pubmed.ncbi.nlm.nih.gov/40561734/

iPSCORE Overview: Induced pluripotent stem cells (iPSCs), derived from human adult cells and capable of being differentiated to become a variety of cell types, are a powerful tool for studying how genetic variants associate with human molecular phenotypes. The Frazer lab has systematically derived and characterized a large unique collection of iPSC lines - The iPSC Collection for Omic Research (iPSCORE). iPSCORE lines are pluripotent with high genomic integrity (no or low numbers of somatic CNVs) as determined using high-throughput RNA-seq and genotyping arrays, respectively.
A total of 273 individuals participated in the study, of which 222 have had iPSCs generated from fibroblasts. Of the 273 individuals, 181 are part of 55 families that include 24 monozygotic twin pairs and 5 dizygotic twin pairs, allowing for the incorporation of familial relationships into genetic analyses. All individuals in iPSCORE have whole genome sequence data.
The Frazer lab has conducted numerous studies using both the iPSCORE iPSC lines and a variety of iPSC-derived cell types including cardiovascular progenitor cells (iPSC-CVPCs), pancreatic precursor cells (iPSC-PPCs), and retina pigment epithelium cells (iPSC-RPEs). We have shown that the iPSCs, CVPCs, and PPCs are suitable surrogate models to identify genetic factors active in early developmental processes because they exhibit fetal-like molecular properties.
The 222 well characterized iPSC lines that constitute the iPSCORE resource are available, please contact Dr. Kelly Frazer if you are interested.
Publications
In total we have published nineteen iPSCORE papers which are listed in chronological order below. In these publications we have addressed a wide range of genetic and biological questions including:
-
how to systematically and effectively reprogram iPSCs from hundreds of individuals and characterize for pluripotency and genome integrity
-
how genetic variants impact gene expression in one of the first large-scale eQTL mapping in iPSCs
-
that non-genetic factors drive aberrant methylation in iPSCs
-
that subclonal mutations, which arise during early reprogramming stages, tend to have large functional impacts compared with clonal mutations
-
that genetic variants impact chromatin loop formation affecting gene expression in iPSCs and CVPCs
-
that iPSC-derived RPE lines can be used to identify and functionally annotate causal variants at AMD risk loci
-
that allele-specific NKX2-5 binding underlies multiple genetic associations with human electrocardiographic traits
-
identified iPSC Gene Signatures and that X Chromosome Dosage are associated with differentiation outcome
-
examined genetic variants in the HLA region in iPSCs discovering that RNF5 expression levels underlie association between the 8.1 ancestral haplotype and colonization in cystic fibrosis
-
described a detailed protocol on how to derive human iPSC-RPEs
-
identified a comprehensive set of structural variants and short tandem repeats
-
examined the properties of structural variants and short tandem repeats associated with gene expression and complex traits
-
described a detailed protocol on how to derive human iPSC-CVPCs
-
examined the spatiotemporal mechanisms of genetic variants underlying cardiac traits and disease
-
demonstrated the regulatory plasticity of eQTLs shared between fetal-like and adult pancreatic tissues
-
identified and characterized regulatory pathways underlying cell state transitions in the iPSCs
-
demonstrated that IFNγ is a potent chromatin remodeler and establishes an IRF-STAT immune-cell like regulatory network
Publications in chronological order
-
 In the first iPSCORE paper we published the initial description of the iPSCORE Resource: Panopoulos AD et al., iPSCORE: A resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types. Stem Cell Reports. 2017 April 6. DOI: 10.1016/j.stemcr.2017.03.012 PMID: 28410642
In the first iPSCORE paper we published the initial description of the iPSCORE Resource: Panopoulos AD et al., iPSCORE: A resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types. Stem Cell Reports. 2017 April 6. DOI: 10.1016/j.stemcr.2017.03.012 PMID: 28410642
Here we described:-
Recruitment and Characterization of Individuals in the iPSCORE Resource
-
Generation, Sample Identity Verification, & Pluripotency Testing of iPSCs
-
Characterization of Somatic Copy-Number Variants using Illumina SNP arrays (HumanCoreExome arrays)
-
Differentiation of iPSC Lines from 3 individuals in iPSCORE family 2 into Cardiomyocytes and Functional Characterization (Time course study – 45 samples)
-
iPSC Lines Carry Genetic Variants Associated with a Variety of Traits and Diseases
-
-
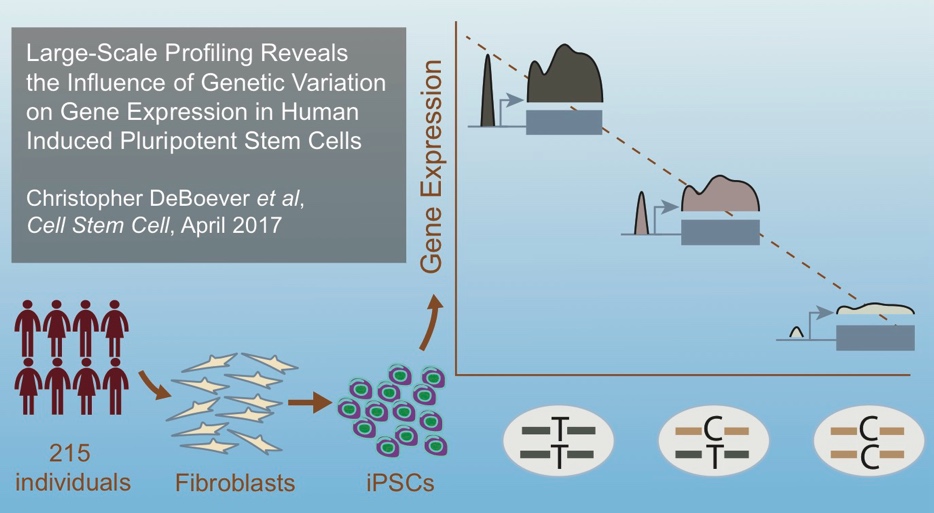 In the second paper we conducted a large-scale iPSC eQTL study: DeBoever C et al., Large-scale profiling reveals the influence of genetic variation on gene expression in human induced pluripotent stem cells. Cell Stem Cell. 2017 April 6. DOI: 10.1016/j.stem. 2017.03.009. PMID: 28388430
In the second paper we conducted a large-scale iPSC eQTL study: DeBoever C et al., Large-scale profiling reveals the influence of genetic variation on gene expression in human induced pluripotent stem cells. Cell Stem Cell. 2017 April 6. DOI: 10.1016/j.stem. 2017.03.009. PMID: 28388430
Here we described:-
eQTL Mapping in 215 iPSC lines from different donors
-
iPSC eQTLs enriched in stem cell regulatory regions
-
Disruption of TF binding sites by eQTL Variants
-
Examined the effect of CNVs on gene expression
-
Examined the effect of rare variants on gene expression
-
X-chromosome reactivation status varies according to gene chromosomal position
-
-
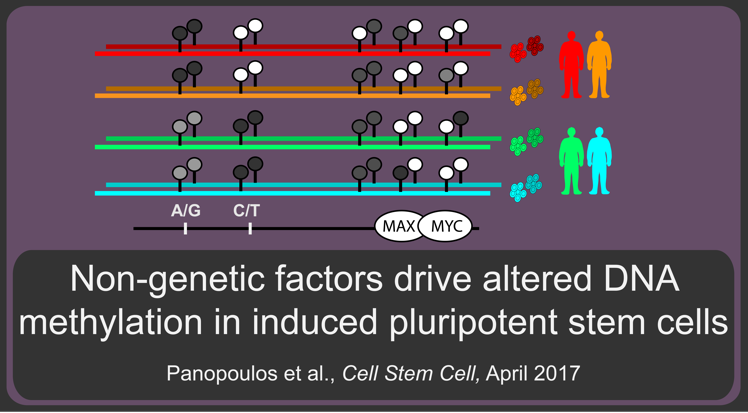 In the third paper we show that non-genetic factors drive aberrant methylation in iPSC lines: Panopoulos AD et al., Aberrant iPSC methylation is associated with motif enrichment and gene expression changes in a clone-specific manner independent of genetics. Cell Stem Cell. 2017 April 6. DOI: 10.1016/j.stem.2017.03.010. PMID: 28388429
In the third paper we show that non-genetic factors drive aberrant methylation in iPSC lines: Panopoulos AD et al., Aberrant iPSC methylation is associated with motif enrichment and gene expression changes in a clone-specific manner independent of genetics. Cell Stem Cell. 2017 April 6. DOI: 10.1016/j.stem.2017.03.010. PMID: 28388429
Here we described:-
Methylation and RNA-Seq profiling of fibroblasts and iPSCs from identical twins
-
Aberrant CpG methylation is recurrent and varies by regulatory region
-
iPSC gain-aberrant CpG sites affect the expression of associated genes
-
Genome-wide association of CpG methylation levels with genetic background, clone, and passage
-
Specific genetic variants near and distal to CpG sites explain the association with genetic background
-
CpG predictor classes are differentially enriched in regulatory regions and for transcription factors
-
Non-genetic factors are associated with methylation and expression of genes relevant for stem cell function
-
Functional characterization of genes enriched for CpG predictor classes
-
iPSC gain-aberrant CpG sites are associated strongly with clone-specific effects
-
Comparison of aberrant genes across studies supports non-genetic mechanisms
-
-
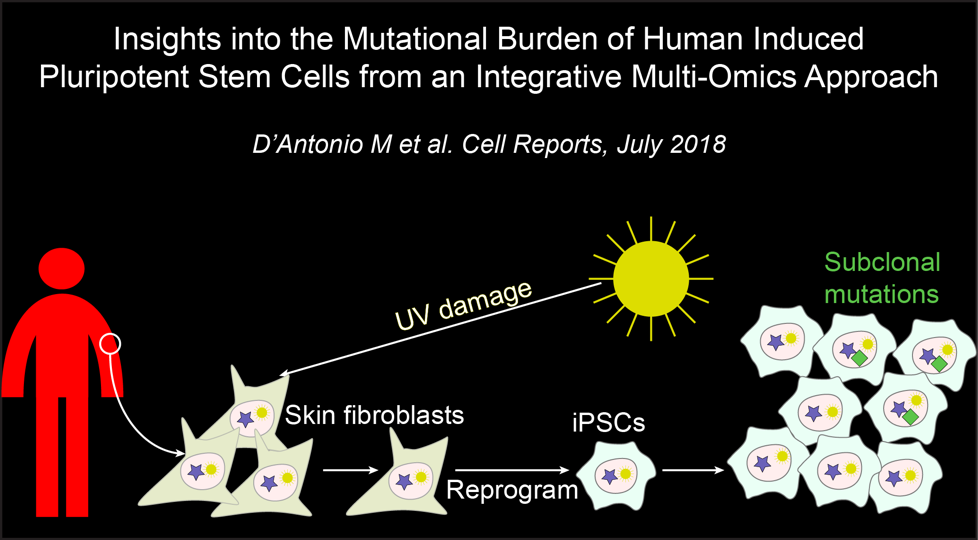 In the fourth paper, we address the mutational burden of human iPSC lines: D’Antonio M et al., Insights into the mutational burden of human induced pluripotent stem cells using an integrative multi-omics approach. Cell Reports. 2018 July 24. doi: 10.1016/j.celrep.2018.06.091. PMID: 30044985.
In the fourth paper, we address the mutational burden of human iPSC lines: D’Antonio M et al., Insights into the mutational burden of human induced pluripotent stem cells using an integrative multi-omics approach. Cell Reports. 2018 July 24. doi: 10.1016/j.celrep.2018.06.091. PMID: 30044985.
Here we described:-
The selection and whole-genome sequencing of 18 iPSC Lines
-
Identification of somatic mutations
-
Comparison of iPSC mutational load to adult stem cells and tumors
-
Evidence of UV damage in the skin-derived iPSC genomes
-
Functional impact of clonal and subclonal mutations on gene structure
-
Clonal and subclonal mutations associated with different chromatin states
-
Subclonal mutations and CNAs result in altered gene expression
-
iPSCs carrying subclonal SNVs do not evolve
-
-
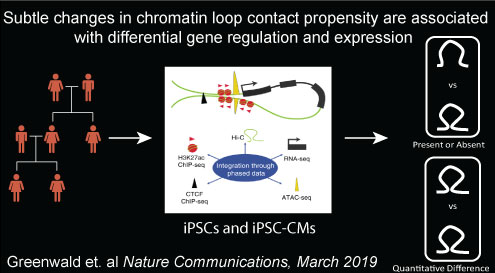 In the fifth paper, we examined how genetic variants impact chromatin loop formation: Greenwald W et al., Subtle changes in chromatin loop contact propensity are associated with differential gene regulation and expression. Nature Communications. 2019 Mar 5. doi: 10.1038/s41467-019-08940-5. PubMed PMID: 30837461.
In the fifth paper, we examined how genetic variants impact chromatin loop formation: Greenwald W et al., Subtle changes in chromatin loop contact propensity are associated with differential gene regulation and expression. Nature Communications. 2019 Mar 5. doi: 10.1038/s41467-019-08940-5. PubMed PMID: 30837461.
Here we described:-
Sample and data collection
-
Identification of chromatin loops in iPSCs and iPSC-CMs
-
Called chromatin loop sets contain a variety of loop types
-
Quantification of differential looping between cell types
-
Cell-type-associated loops (CTALs) are associated with differentiation regulatory changes
-
Functional characterization of CTALs
-
Subtle looping changes are associated with gene regulation
-
Haplotype-based interrogation of loops and gene regulation
-
Haplotype-associated chromatin loops (HTALs) are associated with imprinting and CNVs
-
Regulatory genetic variants and contact propensity
-
Haplotypes, contact propensity, and gene regulation
-
-
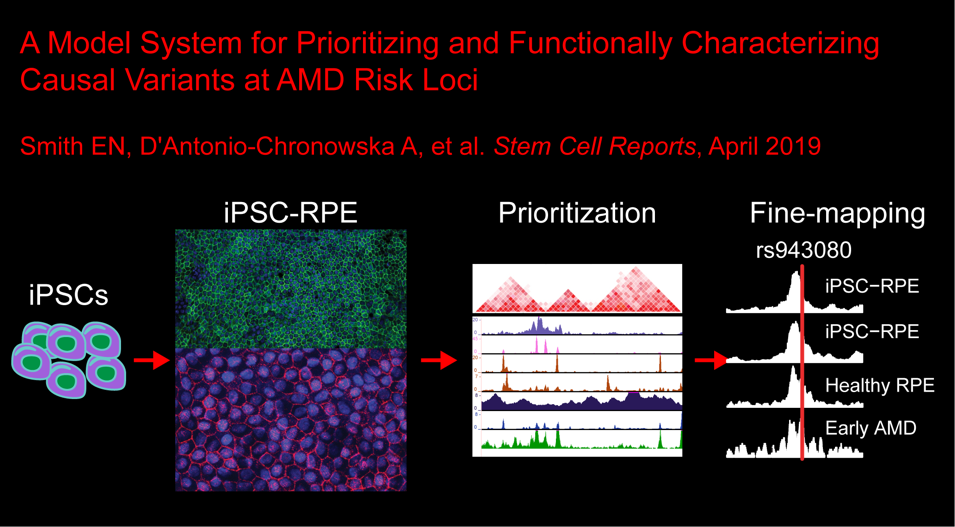 In the sixth paper, we show how we can use iPSC-derived RPE lines to identify and functionally annotate causal variants at AMD risk loci: Smith EN et al., Human iPSC-Derived Retinal Pigment Epithelium: A Model System for Prioritizing and Functionally Characterizing Causal Variants at AMD Risk Loci. Stem Cell Reports. 2019 Apr 25. doi:10.1016/j.stemcr.2019.04.012. PMID: 31080113.
In the sixth paper, we show how we can use iPSC-derived RPE lines to identify and functionally annotate causal variants at AMD risk loci: Smith EN et al., Human iPSC-Derived Retinal Pigment Epithelium: A Model System for Prioritizing and Functionally Characterizing Causal Variants at AMD Risk Loci. Stem Cell Reports. 2019 Apr 25. doi:10.1016/j.stemcr.2019.04.012. PMID: 31080113.
Here we described:-
Derivation of iPSC-RPE
-
The transcriptomes of human iPSC-RPE are similar to those of human fetal RPE
-
Chromatin accessibility of iPSC-RPE
-
Enrichment of regulatory regions for AMD genetic riskFine mapping of AMD-associated loci with fgwas
-
rs943080 is associated with regulatory effects on VEGFA expression
-
-
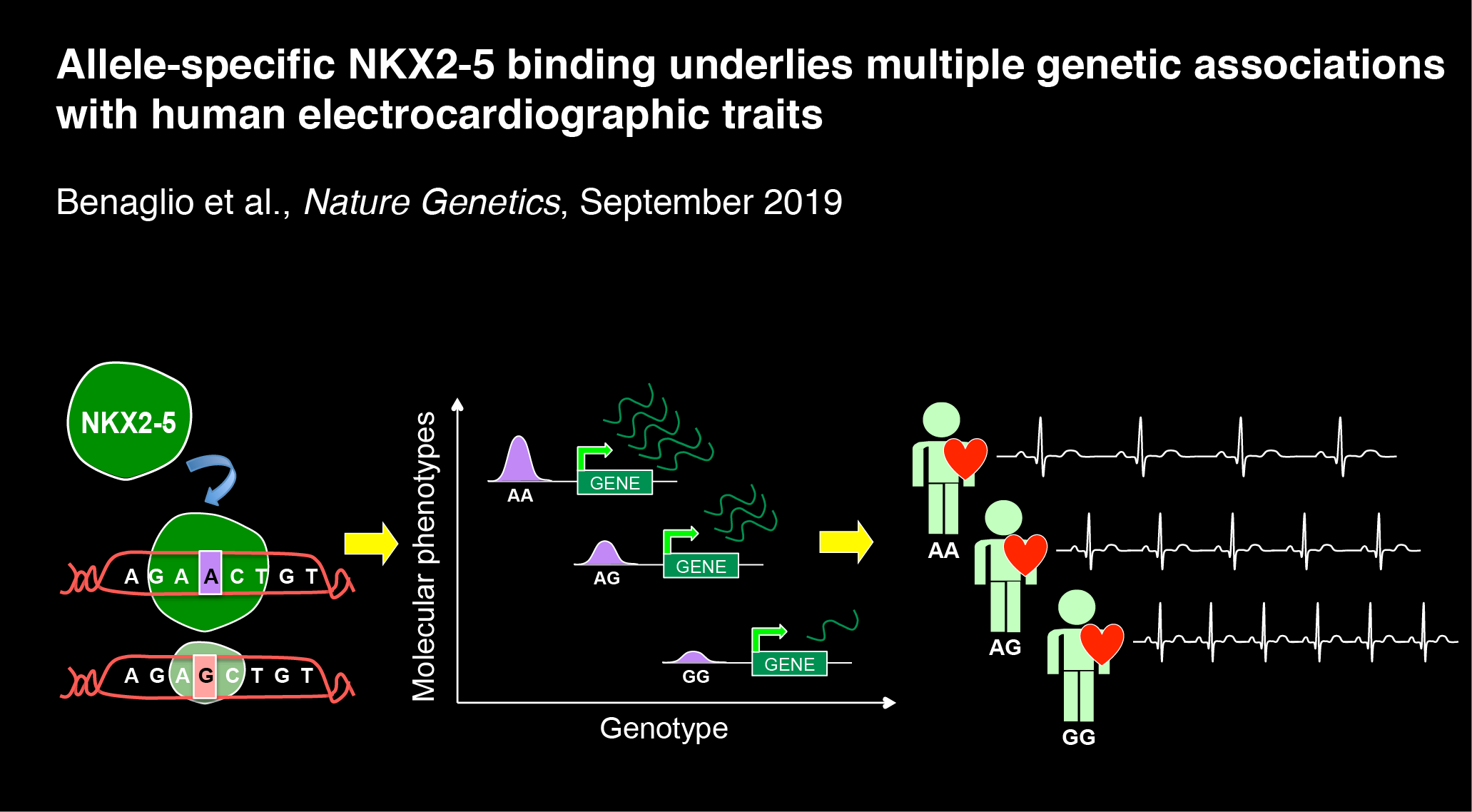 In the seventh paper, we examine how allele-specific binding of NKX2-5 is associated with numerous GWAS loci for electrocardiographic traits: Benaglio P et al., Allele-specific NKX2-5 binding underlies multiple genetic associations with human electrocardiographic traits. Nature Genetics. 2019 Sept 30. doi: 10.1038/s41588-019-0499-3. PMID: 31570892.
In the seventh paper, we examine how allele-specific binding of NKX2-5 is associated with numerous GWAS loci for electrocardiographic traits: Benaglio P et al., Allele-specific NKX2-5 binding underlies multiple genetic associations with human electrocardiographic traits. Nature Genetics. 2019 Sept 30. doi: 10.1038/s41588-019-0499-3. PMID: 31570892.
Here we described:-
Generation and functional genomic profiling of iPSC-derived cardiomyocytes (iPSC-CMs)
-
Genetic background underlies the variability of molecular phenotypes in iPSC-CMs
-
NKX2-5 peaks commonly show allele-specific effects (ASEs)
-
NKX2-5 correlated effects are consistent with dual role as activator and repressor
-
Variation in cardiac TF binding motifs underlies NKX2-5 ASE-SNVs
-
NKX2-5 ASE-SNVs modulate cardiac-specific gene expression
-
NKX2-5 ASE-SNVs are enriched for GWAS associations with EKG traits
-
NKX2-5 ASE-SNVs prioritize causal variants in heart rate GWAS loci
-
Validation of the NKX2-5 ASE-SNV rs3807989 as a functional variant at the CAV1 locus
-
-
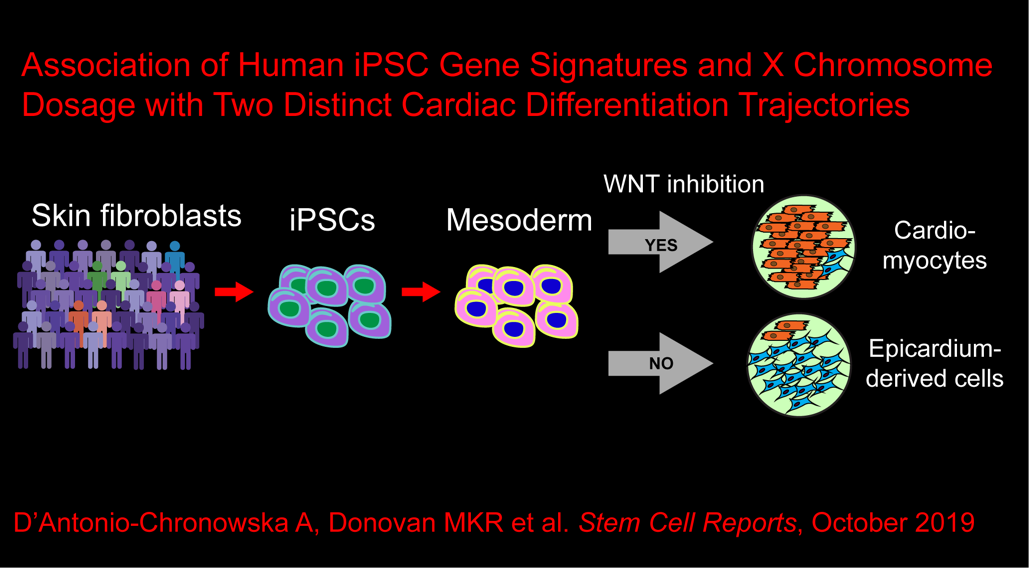 In the eighth paper, we examine how epigenomic features of the iPSCs are associated with the proportions of cell types in iPSC-derived cardiovascular progenitor cells (iPSC-CVPCs): D’Antonio-Chronowska A et al., Association of Human iPSC Gene Signatures and X Chromosome Dosage with Two Distinct Cardiac Differentiation Trajectories. Stem Cell Reports. 2019 October 24. doi: 10.1016/j.stemcr.2019.09.011. PMID: 31668852.
In the eighth paper, we examine how epigenomic features of the iPSCs are associated with the proportions of cell types in iPSC-derived cardiovascular progenitor cells (iPSC-CVPCs): D’Antonio-Chronowska A et al., Association of Human iPSC Gene Signatures and X Chromosome Dosage with Two Distinct Cardiac Differentiation Trajectories. Stem Cell Reports. 2019 October 24. doi: 10.1016/j.stemcr.2019.09.011. PMID: 31668852.
Here we described:-
iPSC-CVPCs Show Cellular Heterogeneity across Samples
-
Subset of Cells Shows Differential Response to WNT Inhibition during Differentiation
-
iPSC-CVPCs Are Composed of Immature CMs and EPDCs
-
iPSC Expression Signatures Affect Cardiac-Fate Differentiation
-
Signature Genes Capture a Large Fraction of the Variance Underlying iPSC Fate Outcome
-
Inherited Genetic Variation Does Not Influence Differentiation Outcome
-
GSEA Implicates ELK1 Targets and Genes on the X Chromosome
-
Sex Is Associated with iPSC Differentiation Outcome
-
Female iPSCs with X Chromosome Reactivation Associated with CM Fate
-
Independent iPSC-CM Derivation Study Validates Findings
-
-
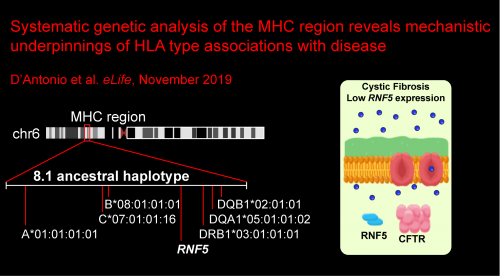 In the ninth paper, we reveal the mechanistic underpinnings of multiple HLA associations with disease. Specifically, we focus on how RNF5 expression levels underlie association between the ancestral HLA haplotype 8.1AH and colonization in cystic fibrosis (CF): D’Antonio M et al., Systematic genetic analysis of the MHC region reveals mechanistic underpinnings of HLA type associations with disease. eLife. 2019 November 20. doi: 10.7554/eLife.48476. PMID: 31746734.
In the ninth paper, we reveal the mechanistic underpinnings of multiple HLA associations with disease. Specifically, we focus on how RNF5 expression levels underlie association between the ancestral HLA haplotype 8.1AH and colonization in cystic fibrosis (CF): D’Antonio M et al., Systematic genetic analysis of the MHC region reveals mechanistic underpinnings of HLA type associations with disease. eLife. 2019 November 20. doi: 10.7554/eLife.48476. PMID: 31746734.
Here we described:-
Eight-digit HLA typing
-
HLA types have high recall rates and reproducibility
-
Eight-digit HLA types associated with expression of cognate HLA gene
-
eQTLs associated with the expression of 56 eGenes in the MHC region
-
Regulatory variants in the MHC region play important roles in complex traits
-
HLA-type eQTLs associated with the expression of 114 eGenes in the MHC region
-
HLA-type eQTLs capture regulatory variation associated with expression of neighboring genes
-
RNF5 expression levels underlie association between 8.1AH and colonization in CF
-
-
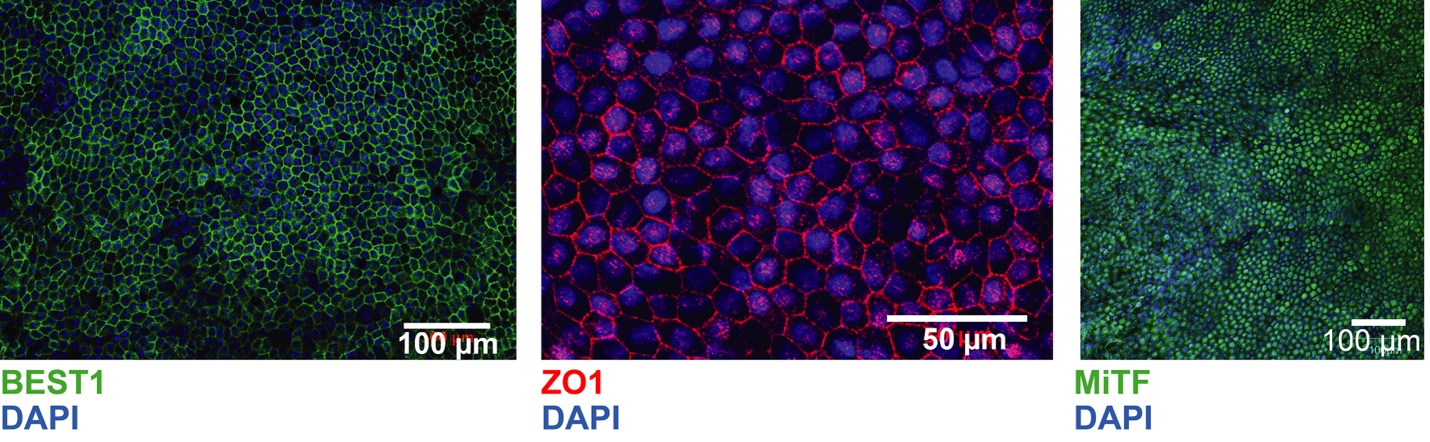 In the tenth paper, we provide a detailed protocol on how to derive human iPSC-RPEs: D’Antonio-Chronowsk A et al., In vitro Differentiation of Human iPSC-derived Retinal Pigment Epithelium Cells (iPSC-RPE). Bio-protocol. 2019 December 20. doi: 10.21769/BioProtoc.3469 PMID: 33654959
In the tenth paper, we provide a detailed protocol on how to derive human iPSC-RPEs: D’Antonio-Chronowsk A et al., In vitro Differentiation of Human iPSC-derived Retinal Pigment Epithelium Cells (iPSC-RPE). Bio-protocol. 2019 December 20. doi: 10.21769/BioProtoc.3469 PMID: 33654959
Our protocol is similar to the protocol of Maruotti et al. (2015), but initiates differentiation when iPSCs reach 80% confluency, which we found to be optimal for all tested iPSCORE lines (Panopoulos et al., 2017). Additionally, we modified the length of time of exposure to small molecules at two steps, which resulted in further increase of yield and purity of iPSC-RPEs (88-98.1% ZO-1 and MiTF double positive cells). Our optimized protocol allowed us to derive iPSC-RPEs from six iPSC lines generated from ethnically diverse individuals under identical culturing conditions without the requirement of any individualized optimization steps. -
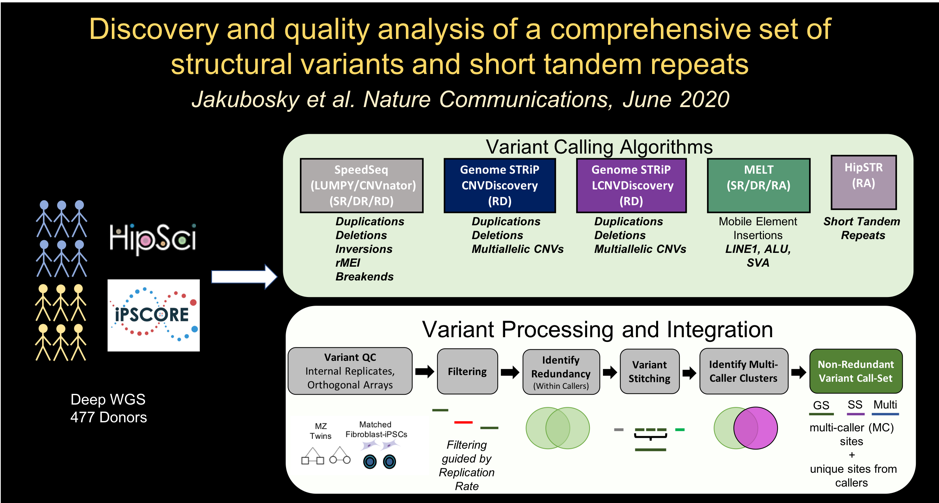 In the eleventh paper, we identify a comprehensive set of structural variants and short tandem repeats: Jakubosky D et al., Discovery and quality analysis of a comprehensive set of structural variants and short tandem repeats. Nature Communications. 2020 June 10. doi: 10.1038/s41467-020-16481-5. PMID: 32522985.
In the eleventh paper, we identify a comprehensive set of structural variants and short tandem repeats: Jakubosky D et al., Discovery and quality analysis of a comprehensive set of structural variants and short tandem repeats. Nature Communications. 2020 June 10. doi: 10.1038/s41467-020-16481-5. PMID: 32522985.
Here we described:-
Comprehensive SV call set
-
Reproducibility of SV calling is associated with quality metrics
-
Creating a high confidence, non-redundant SV call set
-
Variant length, allele frequency, and reproducibility
-
STR genotyping
-
Linkage disequilibrium tagging for SVs and STRs
-
-
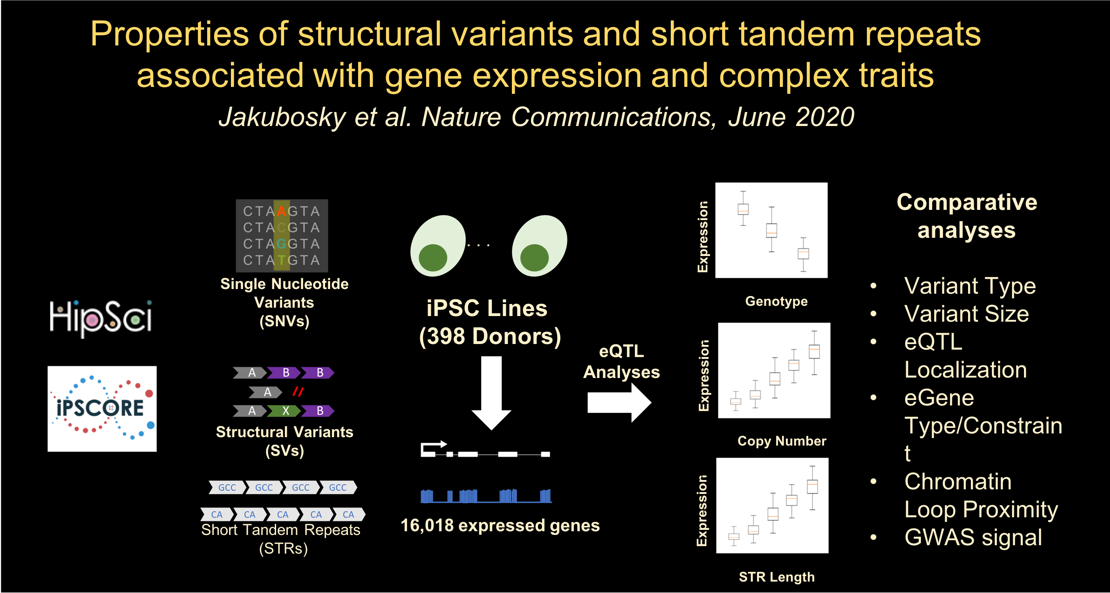 In the twelfth paper, we functionally characterize structural variants and short tandem repeats: Jakubosky D et al., Properties of structural variants and short tandem repeats associated with gene expression and complex traits. Nature Communications. 2020 June 10. doi:10.1038/s41467-020-16482-4. PMID: 32522982.
In the twelfth paper, we functionally characterize structural variants and short tandem repeats: Jakubosky D et al., Properties of structural variants and short tandem repeats associated with gene expression and complex traits. Nature Communications. 2020 June 10. doi:10.1038/s41467-020-16482-4. PMID: 32522982.
Here we described:-
eQTL mapping
-
Copy number variant size influences eQTL associations
-
Multi-allelic CNVs (mCNVs) and deletions are enriched for associations with multiple eGenes
-
Genomic localization of SV and STR eQTLs
-
eVariant type is associated with eGene type and constraint
-
Multi-eGene eQTLs colocalize with distal chromatin loop anchors
-
LD tagging and GWAS associations differ between variant classes
-
-
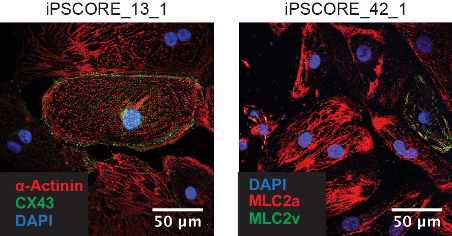 In the thirteenth paper, we provide a detailed protocol on how to derive human iPSC-CVPCs: D’Antonio-Chronowsk A et al., In vitro Differentiation of Human iPSC-derived Cardiovascular Progenitor Cells (iPSC-CVPCs). Bio-protocol 10(18): e3755. DOI: 10.21769/BioProtoc.3755. PMID: 33659414.
In the thirteenth paper, we provide a detailed protocol on how to derive human iPSC-CVPCs: D’Antonio-Chronowsk A et al., In vitro Differentiation of Human iPSC-derived Cardiovascular Progenitor Cells (iPSC-CVPCs). Bio-protocol 10(18): e3755. DOI: 10.21769/BioProtoc.3755. PMID: 33659414.
We have successfully applied this protocol to derive iPSC-CVPCs from 154 different iPSCORE iPSC lines obtaining large quantities of highly pure cardiac cells. An important component of our protocol is cell confluency estimates (ccEstimate), an automated methodology for estimating the time when an iPSC monolayer will reach 80% confluency, which is optimal for initiating iPSC-CVPC derivation, and enables the protocol to be readily used across iPSC lines with different growth rates. Moreover, we showed that cellular heterogeneity across iPSC-CVPCs is due to varying proportions of two distinct cardiac cell types: cardiomyocytes (CMs) and epicardium-derived cells (EPDCs), both of which have been shown to have a critical function in heart regeneration. -
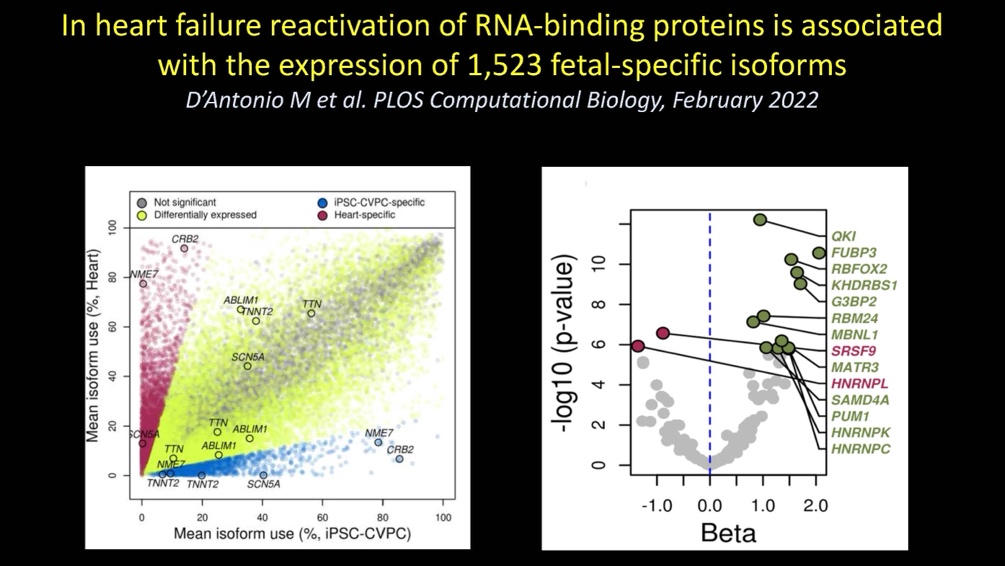 In the fourteenth paper, we examine the role of RNA binding proteins (RBPs) in the expression in heart failure of fetal-specific isoforms that we discovered in the iPSC-CVPCs: D’Antonio M et al., In heart failure reactivation of RNA-binding proteins is associated with the expression of 1,523 fetal-specific isoforms. PLOS Computational Biology. 2022 February 28; doi: 10.1371/journal.pcbi.1009918. PMID: 35226669.
In the fourteenth paper, we examine the role of RNA binding proteins (RBPs) in the expression in heart failure of fetal-specific isoforms that we discovered in the iPSC-CVPCs: D’Antonio M et al., In heart failure reactivation of RNA-binding proteins is associated with the expression of 1,523 fetal-specific isoforms. PLOS Computational Biology. 2022 February 28; doi: 10.1371/journal.pcbi.1009918. PMID: 35226669.
Here we described:-
RNA binding proteins show extensive expression differences between fetal-like and adult cardiovascular system (CVS) tissues
-
Large-scale isoform switching occurs between fetal-like and adult CVS developmental stages
-
iPSC-CVPC-specific exons are enriched for functional protein domains
-
iPSC-CVPC-specific exons are enriched for RBP binding and canonical splice sites
-
Estimated cell type proportions across 966 CVS samples
-
Developmental stage-associated expression differences between fetal-like and adult heart
-
Differential RBP expression associated with developmental stage- and cell type-specific usage of isoforms
-
Heart failure: Reversion to fetal-like RBP and isoform transcriptome
-
-
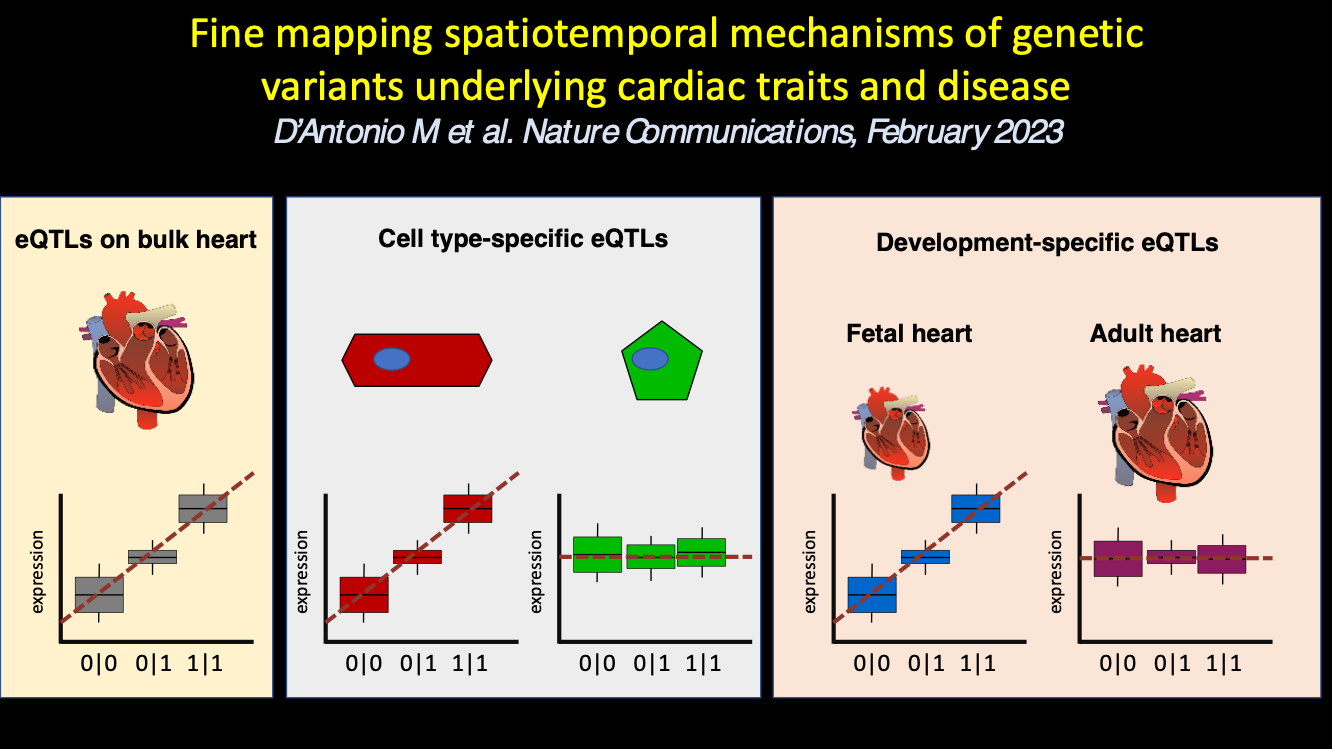 In the fifteenth paper, we examine the iPSC-CVPCs and adult heart tissues to gain insights into the functional context of regulatory variants that are associated with adult cardiac traits and disease: D’Antonio M et al., Fine mapping spatiotemporal mechanisms of genetic variants underlying cardiac traits and disease. Nature Communications. 2023 February 28. doi: 10.1038/s41467-023-36638-2. PMID: 36854752
In the fifteenth paper, we examine the iPSC-CVPCs and adult heart tissues to gain insights into the functional context of regulatory variants that are associated with adult cardiac traits and disease: D’Antonio M et al., Fine mapping spatiotemporal mechanisms of genetic variants underlying cardiac traits and disease. Nature Communications. 2023 February 28. doi: 10.1038/s41467-023-36638-2. PMID: 36854752
Here we described:-
Combined eQTL analysis between fetal-like iPSC-CVPC and adult cardiac tissues in GTEx
-
Mechanisms underlying eQTLs for genes and isoforms iPSC-CVPC-specific exons are enriched for functional protein domains
-
Multigenic eQTL signals are enriched for being spatiotemporal regulated
-
Cardiac genes and their paired antisense RNA enriched for sharing eQTL signals
-
Colocalization identifies potential molecular mechanisms underlying GWAS signals
-
Fine mapping using colocalization data identifies putative causal variants for hundreds of loci
-
Insights from fine mapping into the molecular underpinnings of GWAS signals
-
-
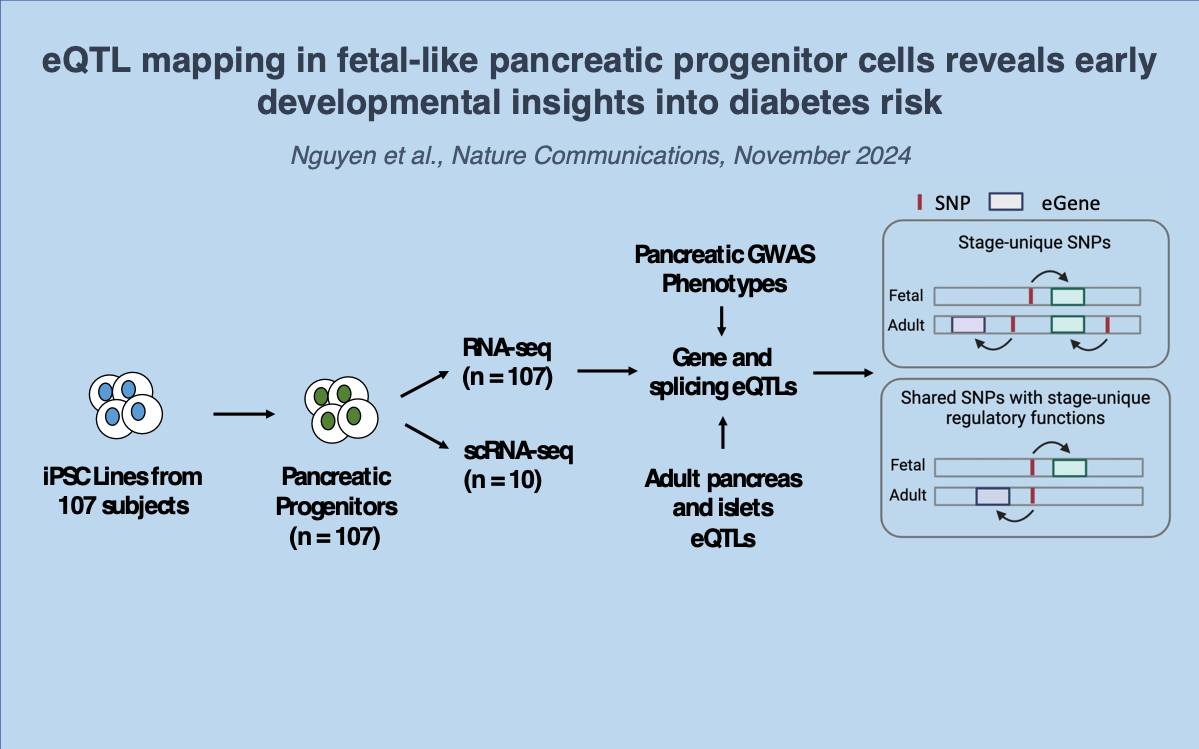 In the sixteenth paper, we examine the iPSC-PPCs and adult pancreas tissues to gain insights into the functional context of regulatory variants contributing to pancreas disease: Nguyen JP et al., eQTL mapping in fetal-like pancreatic progenitor cells reveals early developmental insights into diabetes risk. Nature Communications. 2023 October 30. doi: 10.1038/s41467-023-42560-4. PMID: 37903777
In the sixteenth paper, we examine the iPSC-PPCs and adult pancreas tissues to gain insights into the functional context of regulatory variants contributing to pancreas disease: Nguyen JP et al., eQTL mapping in fetal-like pancreatic progenitor cells reveals early developmental insights into diabetes risk. Nature Communications. 2023 October 30. doi: 10.1038/s41467-023-42560-4. PMID: 37903777
Here we described:-
Large-scale differentiation of fetal-like pancreatic progenitor cells (PPC)
-
Identification and characterization of gene and isoform eQTLs in fetal-like iPSC-PPCs
-
eQTL landscapes of fetal-like iPSC-PPC and adult pancreatic islets
-
Developmental stage-unique and shared eQTLs
-
Regulatory plasticity in eQTLs shared between fetal-like and adult pancreatic tissues
-
Associations of spatiotemporal eQTLs with pancreatic traits and disease phenotypes
-
Spatiotemporally informed eQTL resource provides mechanistic insights into GWAS signals
-
-
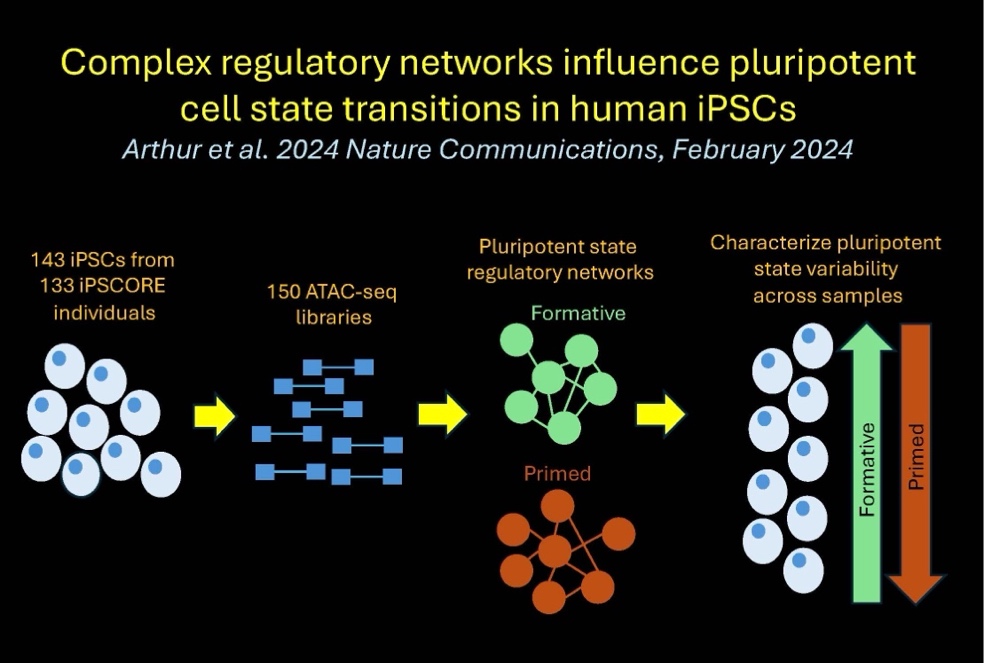 In the seventeenth paper, we identify and characterize regulatory pathways underlying cell state transitions in the iPSCs: Arthur T et al., Complex regulatory networks influence pluripotent cell state transitions in human iPSCs. Nature Communications. 2024 February 23. doi: 10.1038/s41467-024-45506-6. PMID: 38395976
In the seventeenth paper, we identify and characterize regulatory pathways underlying cell state transitions in the iPSCs: Arthur T et al., Complex regulatory networks influence pluripotent cell state transitions in human iPSCs. Nature Communications. 2024 February 23. doi: 10.1038/s41467-024-45506-6. PMID: 38395976
Here we described:-
Relative proportions of pluripotent subpopulations vary across 213 hiPSC lines
-
Identification of gene networks associated with pluripotency state
-
Functional annotation of hiPSC ATAC-seq peaks
-
Functional characterization of regulatory network modules
-
hiPSC regulatory networks are conserved in fetal cell types
-
Characterization of allele-specific chromatin accessibility SNPs (ASCA-SNPs) diseases
-
-
In the eighteenth paper, we show that
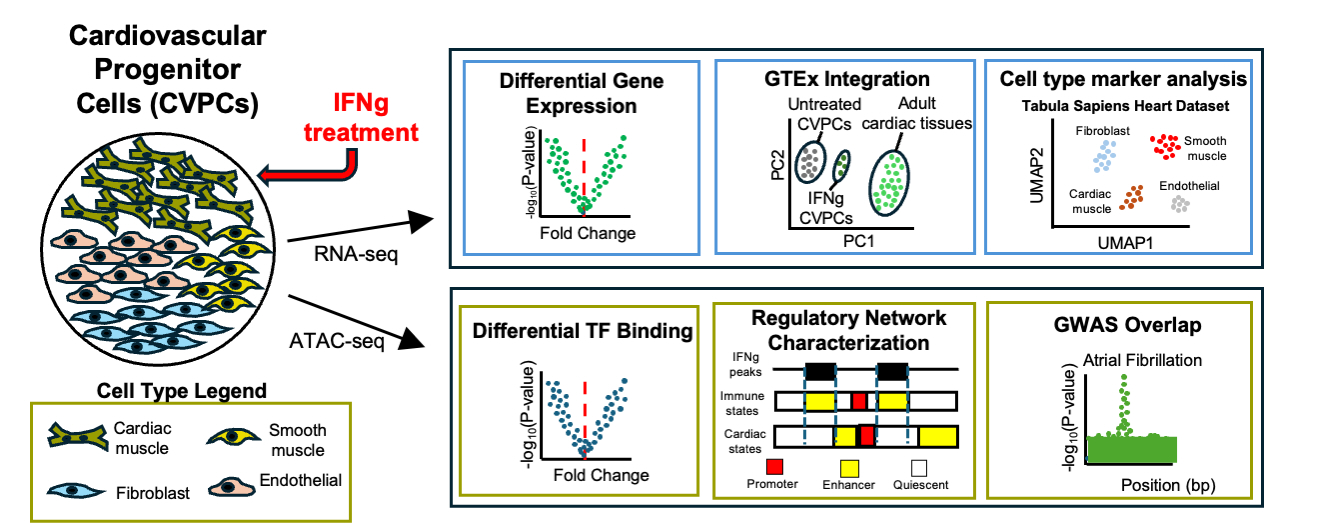 IFNγ induces activation of an immune-like regulatory network in human cardiac tissue and examine the potential role that regulatory elements in this pathway play in common cardiac diseases: Arthur T et al., IFNγ activates an immune-like regulatory network in the cardiac vascular endothelium. Journal of Molecular and Cellular Cardiology Plus. 2025 March 1.
IFNγ induces activation of an immune-like regulatory network in human cardiac tissue and examine the potential role that regulatory elements in this pathway play in common cardiac diseases: Arthur T et al., IFNγ activates an immune-like regulatory network in the cardiac vascular endothelium. Journal of Molecular and Cellular Cardiology Plus. 2025 March 1.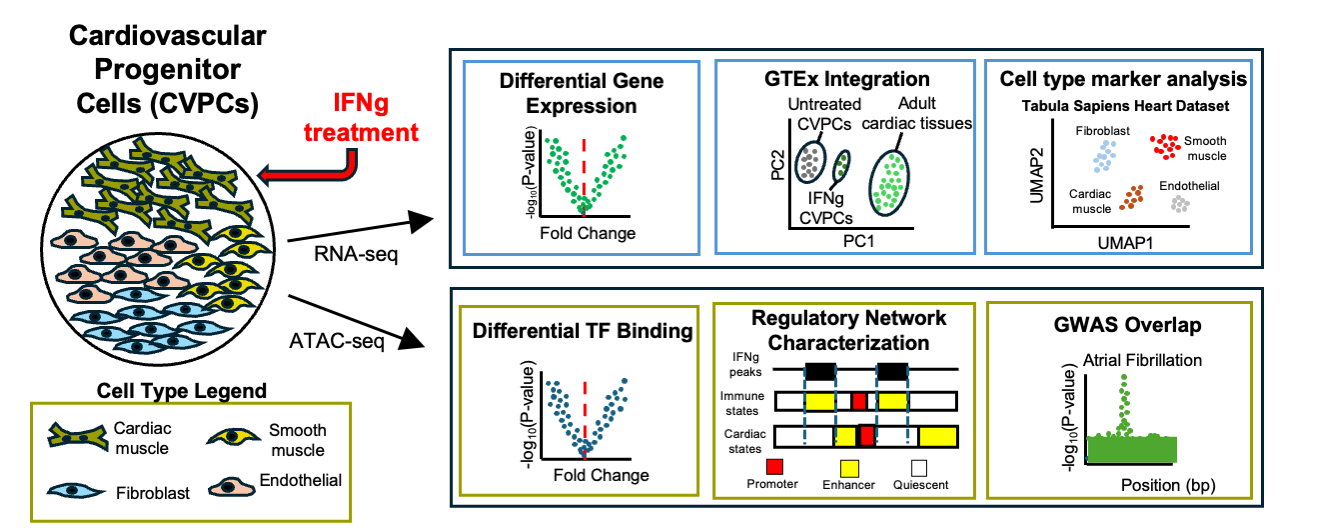 doi: 10.1016/j.jmccpl.2025.100289 PMID: 40104808
doi: 10.1016/j.jmccpl.2025.100289 PMID: 40104808
Here we described:- IFNγ causes significant transcriptomic and epigenomic changes in human cardiac tissue.
- Endothelial population activates an IRF/STAT-mediated immune-like regulatory network.
- GWAS cardiac disease variants are found in IFNγ-induced active chromatin
-
In the nineteenth paper, we utilize the
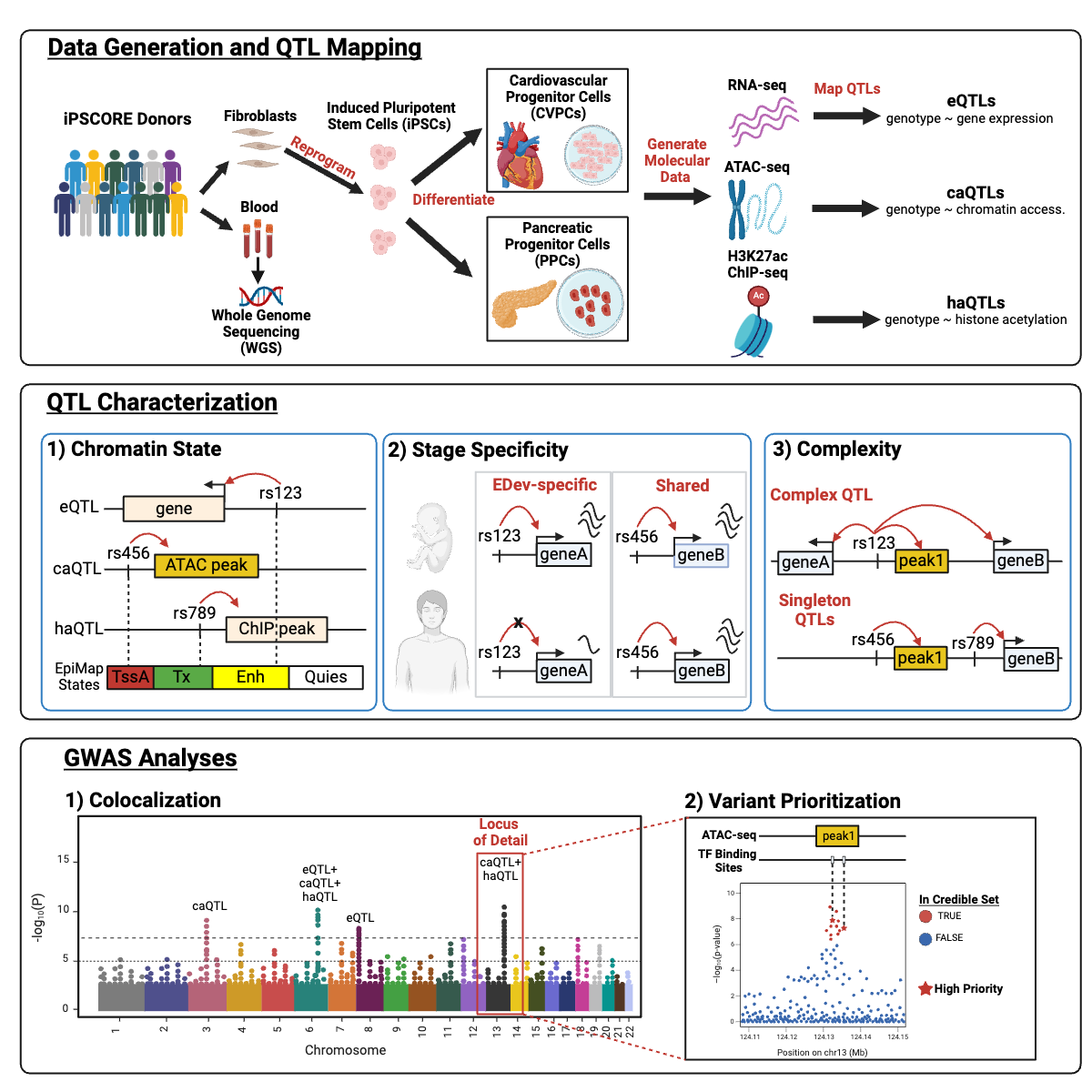 iPSCORE multiomic QTLs to prioritize putative causal variants overlapping transcription factor motifs to elucidate the potential genetic underpinnings of 296 GWAS-QTL colocalizations: Arthur TD and Nguyen JP et al., Multiomic QTL mapping reveals phenotypic complexity of GWAS loci and prioritizes putative causal variants. Cell Genomics 2025 March 12. doi: 10.1016/j.xgen.2025.100775 PMID: 39986281
iPSCORE multiomic QTLs to prioritize putative causal variants overlapping transcription factor motifs to elucidate the potential genetic underpinnings of 296 GWAS-QTL colocalizations: Arthur TD and Nguyen JP et al., Multiomic QTL mapping reveals phenotypic complexity of GWAS loci and prioritizes putative causal variants. Cell Genomics 2025 March 12. doi: 10.1016/j.xgen.2025.100775 PMID: 39986281
Here we described:- Mapped 70,446 multiomic QTLs affecting gene expression and chromatin phenotypes
- Chromatin QTLs capture GWAS loci that are not associated with expression QTLs
- iPSCORE QTLs exhibit temporal activity consistent with early developmental stages
- Multiomic QTLs greatly improve the prioritization of putative causal GWAS variants
- In the twentieth paper, we utilize CRISPR/Cas9
 technology to generate five isogenic human induced pluripotent stem cell (iPSC) lines, one line harboring an LQT1 variant, two lines harboring an LQT3 variant, and two derived control lines: Silva et al., Generation of a set of genetically modified long QT syndrome induced pluripotent stem cell lines carrying knock-in variants rs120074178 (KCNQ1 c.569G > A; p.Arg190Gln) and rs137854600 (SCN5A c.4865G > A; p.Arg1622Gln) and isogenic control lines. Stem Cell Research. 2025 September. doi: 10.1016/j.scr.2025.103755. PMID: 40561734
technology to generate five isogenic human induced pluripotent stem cell (iPSC) lines, one line harboring an LQT1 variant, two lines harboring an LQT3 variant, and two derived control lines: Silva et al., Generation of a set of genetically modified long QT syndrome induced pluripotent stem cell lines carrying knock-in variants rs120074178 (KCNQ1 c.569G > A; p.Arg190Gln) and rs137854600 (SCN5A c.4865G > A; p.Arg1622Gln) and isogenic control lines. Stem Cell Research. 2025 September. doi: 10.1016/j.scr.2025.103755. PMID: 40561734
Here we used an iPSC line from the iPSCORE collection as the parental line to study the genetic and physiological effects arising from missense variants in KCNQ1 and SCN5A genes. The line was derived from dermal fibroblasts of a 25.7 year old Caucasian female (iPSCORE_3_2) using integration-free Sendai virus vectors. Sendai virus clearance and loss of reprogramming genes was confirmed by RT-PCR.